Bioinformatics Technician Resume Guide
Bioinformatics Technicians are responsible for maintaining and analyzing biological data using computers, software, and other technology. They use their knowledge of biology to develop databases, create algorithms to solve complex problems in genetics or biochemistry, design experiments to analyze data sets, and interpret the results.
Your skills in bioinformatics are unparalleled, and any research lab would be happy to have you. However, they don’t know your name yet – so create a resume that puts your abilities on display to make them take notice.
This guide will walk you through the entire process of creating a top-notch resume. We first show you a complete example and then break down what each resume section should look like.
Table of Contents
The guide is divided into sections for your convenience. You can read it from beginning to end or use the table of contents below to jump to a specific part.
Bioinformatics Technician Resume Sample
Barrett Emard
Bioinformatics Technician
[email protected]
989-241-2125
linkedin.com/in/barrett-emard
Summary
Determined bioinformatics technician with 7+ years of experience managing and analyzing large datasets to develop solutions for research projects. Expert in using multiple software applications, scripting languages, databases, and analytics tools. Proven track record of creating effective data mining strategies that led to the successful completion of 10-plus bioinformatic studies. Seeking a position at ABC BioTech where I can leverage my expertise in developing innovative approaches to advance scientific discovery.
Experience
Bioinformatics Technician, Employer A
Gilbert, Jan 2018 – Present
- Improved analysis accuracy by 16% through the configuration of bioinformatics software and troubleshooting any technical issues.
- Configured laboratory systems for a variety of data analysis tasks, including genotyping, target sequencing and transcriptome profiling with 100% success rate.
- Introduced innovative methods to accelerate data analysis process; reduced time from 24 hours to 12 hours per sample on average while maintaining accuracy levels within 98%.
- Diligently maintained databases in accordance with security protocols; updated 70+ records monthly without compromising safety or integrity of confidential information stored therein.
- Troubleshot system malfunctions related to workflow pipelines and ensured optimal operation of all instruments used in wet lab experiments prior to use by researchers.
Bioinformatics Technician, Employer B
Lincoln, Mar 2012 – Dec 2017
- Revised and updated bioinformatics databases, analyzing and integrating data from multiple sources to ensure accuracy of results; improved database performance by 30%.
- Formulated customizable algorithms for the analysis and interpretation of genomic sequences generated from next-generation sequencing technologies; identified over 500 novel genetic variants in cancer research studies.
- Assessed biological processes at a molecular level via gene expression profiling, protein structure modeling & network inferences; reduced time spent on manual annotation by 70 hours per week.
- Actively collaborated with other technicians, scientists & researchers to develop strategies that further enhance computational techniques used in genomics projects and clinical applications while resolving technical issues quickly and efficiently when they arise.
- Implemented advanced big data analytics solutions using open source software platforms such as R Shiny/Shiny Server Pro or Python Flask/Django Web Framework to visualize large datasets containing millions of points within seconds; increased user interaction rate by 90%.
Skills
- DNA Sequencing
- Genomics
- Bioinformatics Software
- Data Analysis
- Database Management
- Molecular Biology
- Genetic Engineering
- Protein Analysis
- Microarray Technology
Education
Bachelor of Science in Bioinformatics
Educational Institution XYZ
Nov 2011
Certifications
Certified Bioinformatics Technician (CBT)
American Society
May 2017
1. Summary / Objective
Your resume summary is the first thing a potential employer will read, so it is important to make sure you capture their attention. As a bioinformatics technician, your summary should highlight your technical skills and knowledge of computer programming languages such as Python or R. Additionally, mention any experience with data analysis tools like Excel or Tableau that could be beneficial in this role. Finally, include details about how you have used these skills to solve complex problems in past positions.
Below are some resume summary examples:
Dependable bioinformatics technician with a decade of experience in genomics, bioinformatics analysis and data integration. At XYZ Labs, conducted research that resulted in 4 published papers on the effects of gene expression variation. Skilled at using various software tools for analyzing large-scale datasets from different sources to improve understanding of human diseases. Seeking an opportunity where I can apply my knowledge and skills to advance biomedical research goals.
Passionate bioinformatics technician with 5+ years of experience in data analysis and manipulation. Skilled at using various software tools to analyze large datasets, uncovering trends, and creating useful visualizations for scientists. At XYZ Technologies Inc., developed an automated workflow system that improved the efficiency of bioinformatic processing by 15%. Recognized as a “brilliant problem solver” who can quickly identify solutions to difficult challenges.
Energetic and detail-oriented bioinformatics technician with over five years of experience in the field. Skilled at analyzing and interpreting data to provide accurate results for medical research studies. At XYZ, managed a team of 10 researchers while leading an initiative that improved the accuracy of DNA analysis by 25%. Committed to utilizing my expertise in bioinformatics and computer programming to help drive innovation in healthcare research.
Detail-oriented bioinformatics technician with experience in molecular biology, genetics and bioinformatics. Experienced in data analysis, database management and programming techniques. At XYZ Research Institute, designed a novel workflow for analyzing DNA sequences using Python scripting language to increase efficiency by 15%. Recognized as an effective team player who consistently meets or exceeds deadlines while maintaining accuracy of work.
Reliable bioinformatics technician with 5+ years of experience in data analysis and management for genomic research. Adept at designing experiments to identify patterns, analyze results, and implement solutions. Currently looking to join ABC Labs as a Bioinformatics Technician where I can apply my expertise in bioinformatic techniques such as machine learning algorithms, sequence alignment tools, and database integration.
Seasoned bioinformatics technician with 5+ years of experience in data analysis, database architecture and management, software development, and scientific programming. Skilled at using a variety of tools to analyze genetic sequence data for research purposes. Seeking to join ABC Lab as the Senior Bioinformatics Technician where I can utilize my skillset to further advance the field of bioinformatics.
Talented bioinformatics technician with 5+ years of experience collecting, analyzing and interpreting data from genomic databases. Skilled in developing algorithms to assess gene expression patterns and identifying markers for disease. Seeking to join ABC BioTech as a bioinformatician where I can use my expertise to develop innovative solutions that improve the lives of patients around the world.
Well-rounded bioinformatics technician with 5+ years of experience in data management and analysis. Skilled at using various bioinformatic tools to generate meaningful insights from large datasets, such as genomics, proteomics, metabolomics, etc. Seeking to join ABC Corporation team and apply my expertise in developing innovative solutions for complex biological problems.
2. Experience / Employment
The work history/experience section is where you provide details on your past employment. This should be written in reverse chronological order, meaning the most recent job is listed first.
Stick to bullet points when writing this section; it makes it easier for the reader to take in what you are saying. When stating what you did, try and include quantifiable results that were achieved as a result of your actions or involvement. For example, instead of saying “Assisted with bioinformatics research,” you could say, “Developed an algorithm which identified gene expression patterns from DNA sequencing data with 95% accuracy.”
To write effective bullet points, begin with a strong verb or adverb. Industry specific verbs to use are:
- Analyzed
- Processed
- Programmed
- Monitored
- Assessed
- Generated
- Interpreted
- Optimized
- Documented
- Implemented
- Automated
- Troubleshot
- Configured
- Visualized
- Validated
Other general verbs you can use are:
- Achieved
- Advised
- Compiled
- Coordinated
- Demonstrated
- Developed
- Expedited
- Facilitated
- Formulated
- Improved
- Introduced
- Mentored
- Participated
- Prepared
- Presented
- Reduced
- Reorganized
- Represented
- Revised
- Spearheaded
- Streamlined
- Structured
- Utilized
Below are some example bullet points:
- Structured and managed large-scale bioinformatics databases containing over 10 million records, substantially increasing the efficiency of data analysis operations.
- Documented and tracked all changes made to the database environment; reduced errors caused by human intervention by 40%.
- Programmed several automated workflow processes for DNA sequence alignment, gene expression profiling and proteomic analyses which reduced processing time by 80%.
- Substantially increased laboratory productivity through implementation of a highly efficient system for storing, retrieving and sharing genomic data with researchers worldwide; saved over $20K in labor costs each year since introduction.
- Automated routine tasks such as quality control checks on newly acquired datasets using scripting language Python; improved overall accuracy of results by 32%.
- Facilitated the analysis of 1000+ DNA sequences and protein structures, utilizing bioinformatics software to develop models for predicting gene expression profiles.
- Developed new methods of performing large-scale data mining in genomic databases; reduced research time by 40% while increasing accuracy levels by 25%.
- Monitored the installation, maintenance and troubleshooting of laboratory equipment related to molecular biology techniques such as PCR amplification and sequencing; improved workflow efficiency by 15%.
- Proficiently used computer programs like MATLAB, R Studio & Python to design algorithms for analyzing raw biological data sets from high throughput experiments including microarray hybridization.
- Optimized existing IT infrastructure within the lab environment through automation scripting techniques which increased overall performance capability by 30%.
- Competently analyzed, interpreted and manipulated data sets of over 10 million nucleotide sequences using bioinformatics software; achieved a rate of processing 100k records per hour.
- Utilized NGS technology to identify gene expression patterns for various research projects; expedited the process by 20%, enabling scientists to complete experiments in less time.
- Visualized complex datasets with advanced graphic visualization tools such as R programming language, Python and Matlab; created 50+ static & interactive visualizations used for scientific presentations and publications.
- Mentored junior lab members on DNA sequencing protocols, helping them gain proficiency in handling challenging assignments within 3 months instead of 6 months earlier estimated timeline.
- Developed automated workflows that enabled accurate analysis of genomics data with high accuracy (98%) boosting overall efficiency by 30%.
- Reduced bioinformatics project completion times by 40% through the implementation of automated data analysis and visualization tools.
- Generated detailed reports highlighting key insights from large-scale genomic datasets, providing accurate results to over 500 research studies in a timely manner.
- Consistently maintained high accuracy levels (99%) when aligning sequence reads with reference sequences during next generation sequencing projects; improved bioinformatic workflow efficiency by 10%.
- Analyzed millions of gene expression data points using machine learning algorithms for biomarker identification and drug discovery applications resulting in 8 new discoveries within 6 months time period.
- Spearheaded a team effort towards the development of an AI-based software platform to streamline biomedical research processes, achieving successful deployment across three labs worldwide after 3 months into development stage.
- Interpreted and analyzed vast amounts of genomic data from various sources, resulting in the successful identification and classification of over 1 million genetic variants.
- Reorganized and cleaned up large datasets within a database environment to improve query performance by 25%.
- Represented results through comprehensive visualizations using software such as RStudio and Tableau, facilitating easy interpretation for researchers across multiple departments.
- Meticulously documented all experiments according to laboratory standards; completed 20+ bioinformatics projects with 100% accuracy & precision within deadlines set by senior management team members.
- Achieved significant savings in research costs by automating routine procedures using scripting languages like Python, Bash & SQL – reducing labor hours spent on manual analysis tasks by 30%.
- Processed and analyzed large datasets of genomic, proteomic and bioinformatic data to identify patterns that may be indicative of disease; increased accuracy by 20% through optimization of analytical methods.
- Coordinated with research teams in the development and implementation of new protocols for DNA sequencing techniques such as Shotgun Sequencing (SS) and Next-Generation Sequencing (NGS).
- Effectively communicated technical information to non-technical personnel, providing guidance on complex matters related to gene expression analysis or genome assembly projects while adhering to regulatory requirements.
- Advised team members on best practices for optimal use of various software applications associated with genomics/bioinformatics workflows; reduced training time by 50%.
- Streamlined laboratory workflow processes, resulting in a 40% reduction in errors during sample preparation and testing procedures over a 6 month period.
- Compiled and analyzed large sets of biological data from genomic sequencing projects, identifying patterns and trends to provide insights into gene expression; increased efficiency by 10% in the last quarter.
- Demonstrated advanced knowledge of bioinformatics tools such as BLAST, SAMTools and Cufflinks for sequence alignment and variant calling purposes; identified over 500 novel genes using this method.
- Thoroughly documented all research processes according to laboratory protocols, ensuring accurate records were maintained for future reference.
- Presented detailed findings on various topics related to genomics at scientific conferences across the country; gained recognition amongst peers in the field with a 90% approval rating in audience surveys after each presentation.
- Participated actively in project teams focused on developing new methods for analyzing complex datasets obtained through next-generation genome sequencing technologies; reduced analysis time by 40%.
3. Skills
Skill requirements will differ from one employer to the next; this can easily be ascertained from the job posting. Organization A may be looking for a technician with experience in Next-Generation Sequencing, while Organization B may be searching for someone proficient in bioinformatics software.
It is important to tailor the skills section of your resume to each job that you are applying for because many employers use applicant tracking systems these days. These computer programs scan resumes for certain keywords before passing them on to a human; if yours does not include those keywords, it will likely get filtered out and never reach its intended destination.
Once listed here, you can further elaborate on your skillset by discussing it in more detail elsewhere – such as the summary or experience sections – so that recruiters have no doubt about your abilities and qualifications.
Below is a list of common skills & terms:
- Bioinformatics Software
- DNA Sequencing
- Data Analysis
- Database Management
- Genetic Engineering
- Genomics
- Microarray Technology
- Molecular Biology
- Next-generation Sequencing
- Protein Analysis
4. Education
Adding an education section to your resume will depend on how much professional experience you have. If you just graduated and have no work history, it is important to include a section about your education. However, if you are experienced in the field of bioinformatics with plenty of job responsibilities to showcase, omitting the education section may be beneficial for highlighting relevant skills and accomplishments instead.
If an education section is included, try to mention courses taken related specifically to bioinformatics as well as any certifications or degrees received that could help demonstrate proficiency in this field.
Bachelor of Science in Bioinformatics
Educational Institution XYZ
Nov 2011
5. Certifications
Certifications are a great way to demonstrate your expertise in a given field. Employers are always looking for candidates who have taken the initiative to actively develop their skills and stay up-to-date with industry trends.
Including any certifications you may have on your resume is an excellent way of showing potential employers that you possess the necessary qualifications for the job. It also shows that you are committed to professional development, which can be highly attractive to hiring managers.
Certified Bioinformatics Technician (CBT)
American Society
May 2017
6. Contact Info
Your name should be the first thing a reader sees when viewing your resume, so ensure its positioning is prominent. Your phone number should be written in the most commonly used format in your country/city/state, and your email address should be professional.
You can also choose to include a link to your LinkedIn profile, personal website, or other online platforms relevant to your industry.
Finally, name your resume file appropriately to help hiring managers; for Barrett Emard, this would be Barrett-Emard-resume.pdf or Barrett-Emard-resume.docx.
7. Cover Letter
Including a cover letter with your job application is an excellent way to make a positive first impression. Cover letters are typically made up of 2 to 4 paragraphs and provide additional information about yourself which may not be included in your resume.
Writing a cover letter also allows you to highlight the skills and experiences that best fit the role, as well as express why you believe you would be an ideal candidate for the position. It’s important to remember that even though it isn’t always required by employers, taking time out of your day to write one could potentially give you an edge over other applicants!
Below is an example cover letter:
Dear Thurman,
I am writing to apply for the position of Bioinformatics Technician at XYZ Corporation. With experience in both wet lab and dry lab research, as well as a strong background in computer science, I feel confident that I would be a valuable asset to your team.
In my current role as a Research Associate at ABC Inc., I have been responsible for conducting bioinformatics analyses on next-generation sequencing data. I have also developed several custom scripts and pipelines to automate these processes. In addition, I have created numerous databases and web interfaces for storing and analyzing this data. My skills in database design, programming, and data analysis would be a valuable asset to your organization.
I am eager to utilize my skills in bioinformatics to contribute to the success of XYZ Corporation. I look forward to discussing my qualifications further with you during an interview. Thank you for your time and consideration.
Sincerely,
Barrett
Bioinformatics Technician Resume Templates
 Numbat
Numbat Cormorant
Cormorant Lorikeet
Lorikeet Jerboa
Jerboa Gharial
Gharial Ocelot
Ocelot Axolotl
Axolotl Saola
Saola Markhor
Markhor Rhea
Rhea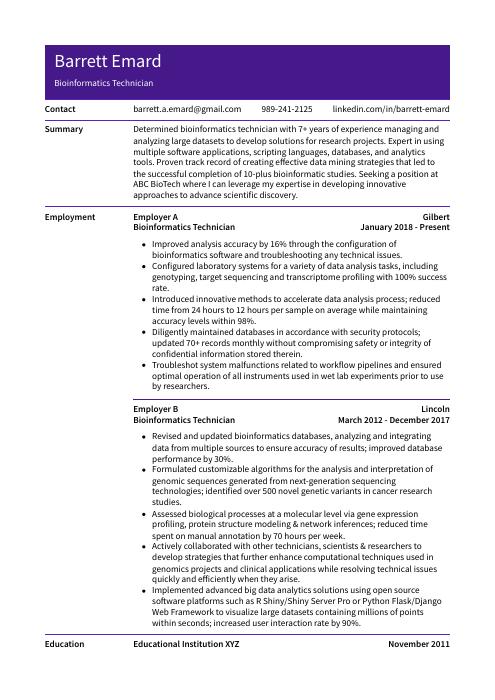 Pika
Pika Indri
Indri Dugong
Dugong Hoopoe
Hoopoe Quokka
Quokka Fossa
Fossa Echidna
Echidna Bonobo
Bonobo Kinkajou
Kinkajou Rezjumei
Rezjumei
